本次澳洲代写是一个生物相关的实验报告
Design, building and testing of cellular biosensors for heavy metals pollutants
3. Online Practical Week 2 – Assembly of biosensors
Objective: To assemble individual DNA parts into cell-based biosensors for detecting heavy metal contamination. The DNA parts will be combined using an automatic pipetting robot and assembled into circular plasmid DNA using Gibson assembly (described in Lecture 2). The resulting plasmid DNA molecules will be subsequently transformed into competent E. coli cells, and the cells plated onto antibiotic selection plates.
Materials and Equipment
• DNA parts (see below)
• Gibson enzyme master mix
• OT-2 liquid handler robot (shown in more detail during the lab practical)
• PCR and 1.5 mL tubes
• PCR thermocycler
• Ice in an esky and hot water bath set to 42°C
• Competent E. coli cells
• SOC media
• LB-agar + kanamycin plates
• Pipettes and sterile tips
• Disposable plate spreaders
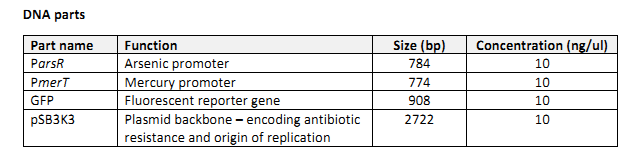
The DNA parts were prepared either by using PCR to amplify a DNA template contained within a
plasmid (the promoters and reporter genes) or by linearising a circular plasmid with restriction
enzymes (the pSB3K3 plasmid backbone). Several subsequent steps were performed to remove any remaining plasmids from the samples, leaving only the linear DNA parts to be mixed and assembled.
Gibson assembly of DNA parts
Gibson assembly is a molecular cloning method for joining multiple DNA fragments into a single molecule. It has been rapidly adopted by the synthetic biology community due to its ease-of-use, flexibility and suitability for assembly of large DNA constructs. This method can be used to assemble multiple DNA molecules, even entire plasmids, up to many hundreds of kilobases in length. Refer to lecture 2 for a complete description of how the Gibson assembly reaction works.
The Gibson assembly will be used to assemble a particular promoter and reporter gene with the
pSB3K3 plasmid DNA backbone. The pSB3K3 plasmid DNA contains a kanamycin antibiotic resistance gene and an origin of replication to enable propagation of the assembled plasmid in a bacterial host.
The reaction is usually preformed with a Gibson assembly master mix that contains three enzymes and their required substrates. To perform the assembly, the researcher simply combines the DNA fragments with appropriate overhangs and the master mix, followed by an incubation step.
A critical requirement of the Gibson assembly is that there must be similar numbers of the DNA
molecules of each fragment to be joined. Therefore, you would normally add equimolar amounts of each DNA component to the reaction mix.
In this practical, however, we will use approximations to prepare the Gibson reaction and avoid
pipetting complicated volumes. In this example, the arsenite and mercury promoters and GFP parts are approximately 1 kb in length and the linearised backbone is approximately 3 kb in length. Since the backbone is roughly three times longer than the other parts, it is necessary to add three times the mass (ng) of the backbone, relative to the other parts, to obtain the same quantity (pmol).
This approximation works well for most Gibson reactions.